
Optimization and Computational Systems Biology Lab
|
SOFTWARES
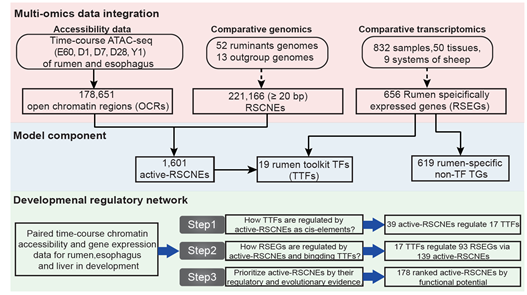
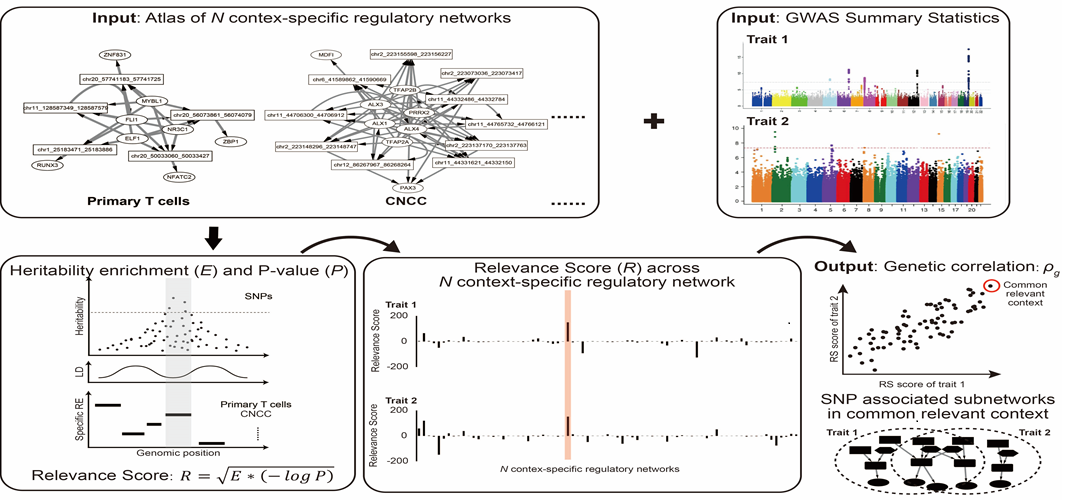
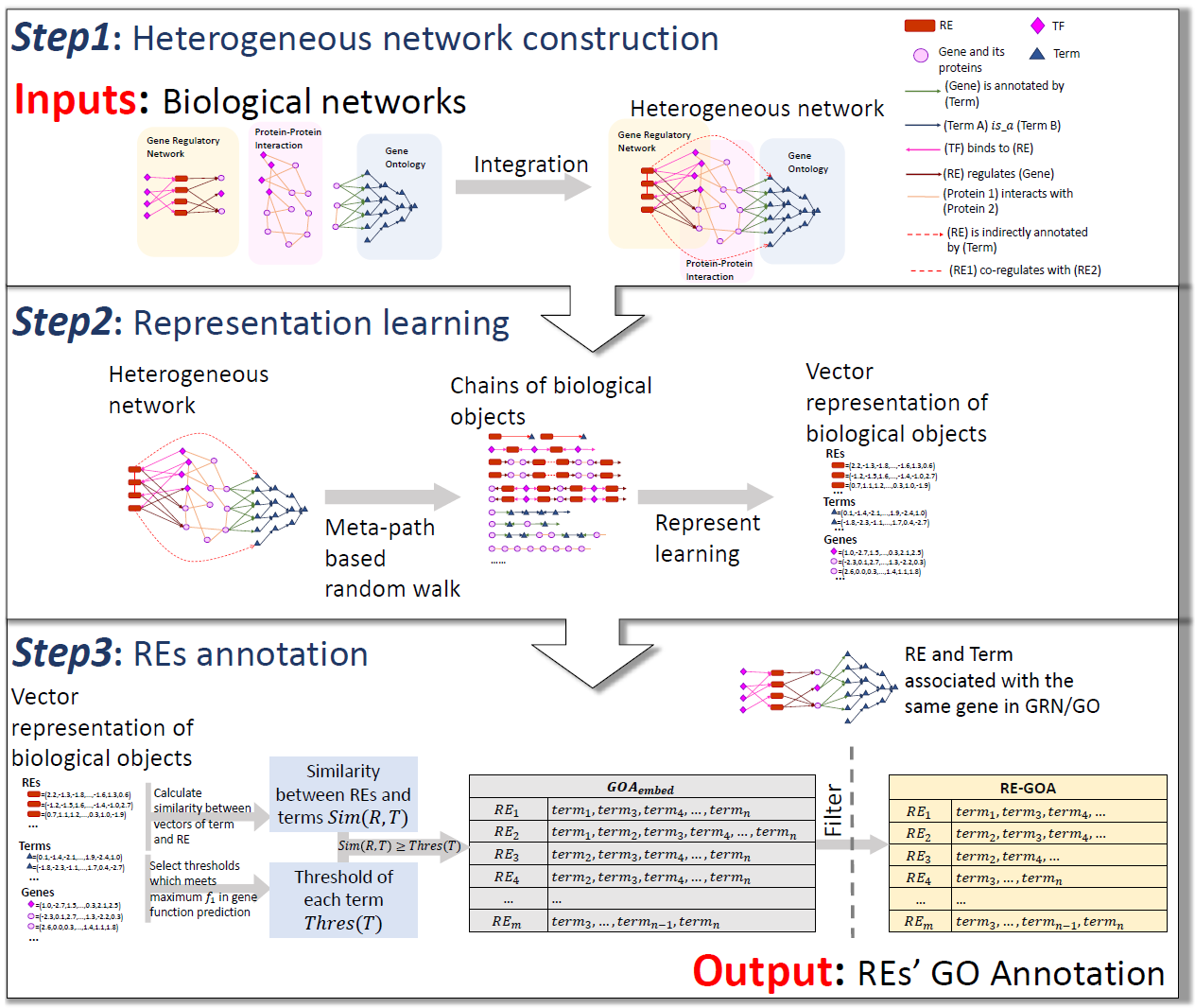
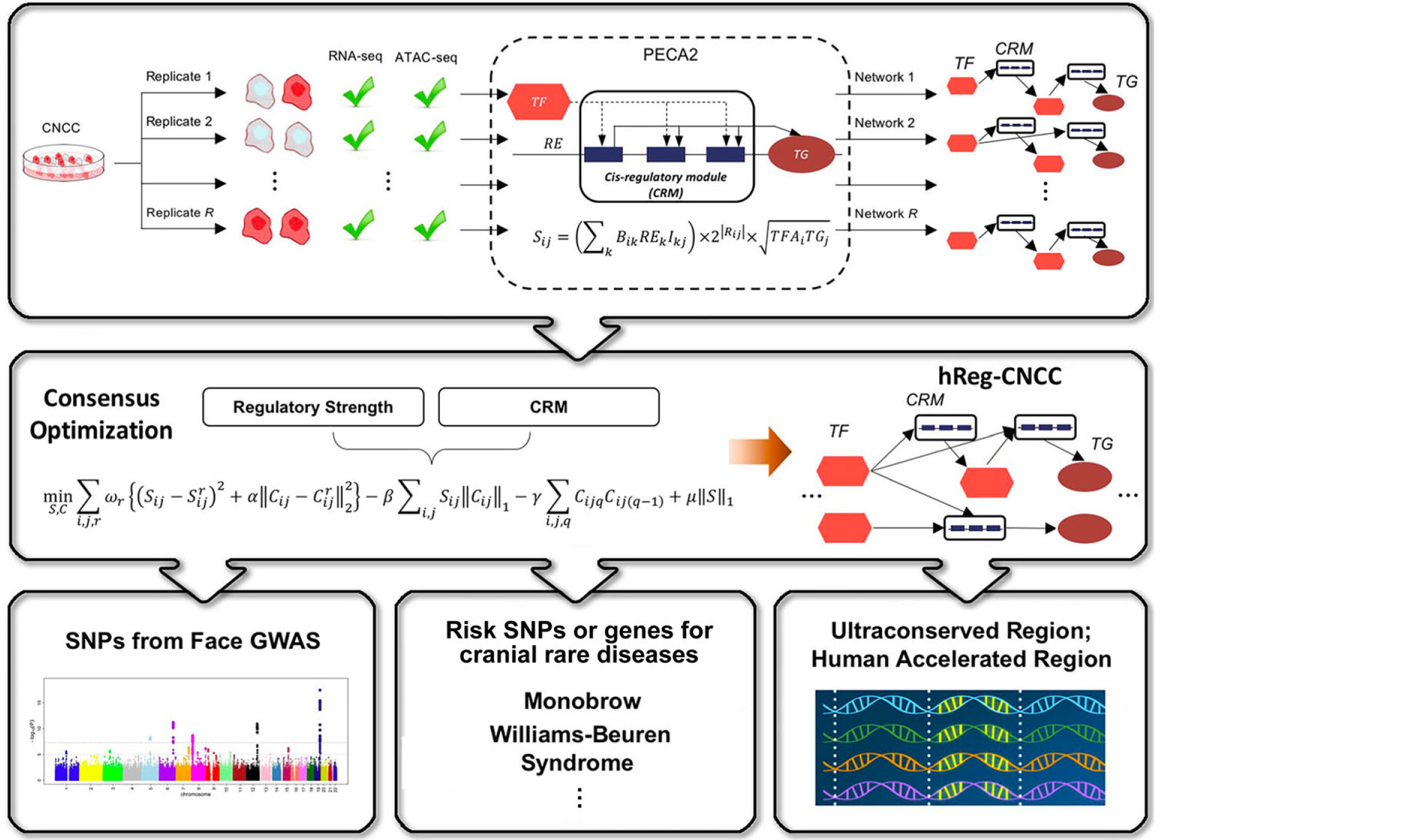
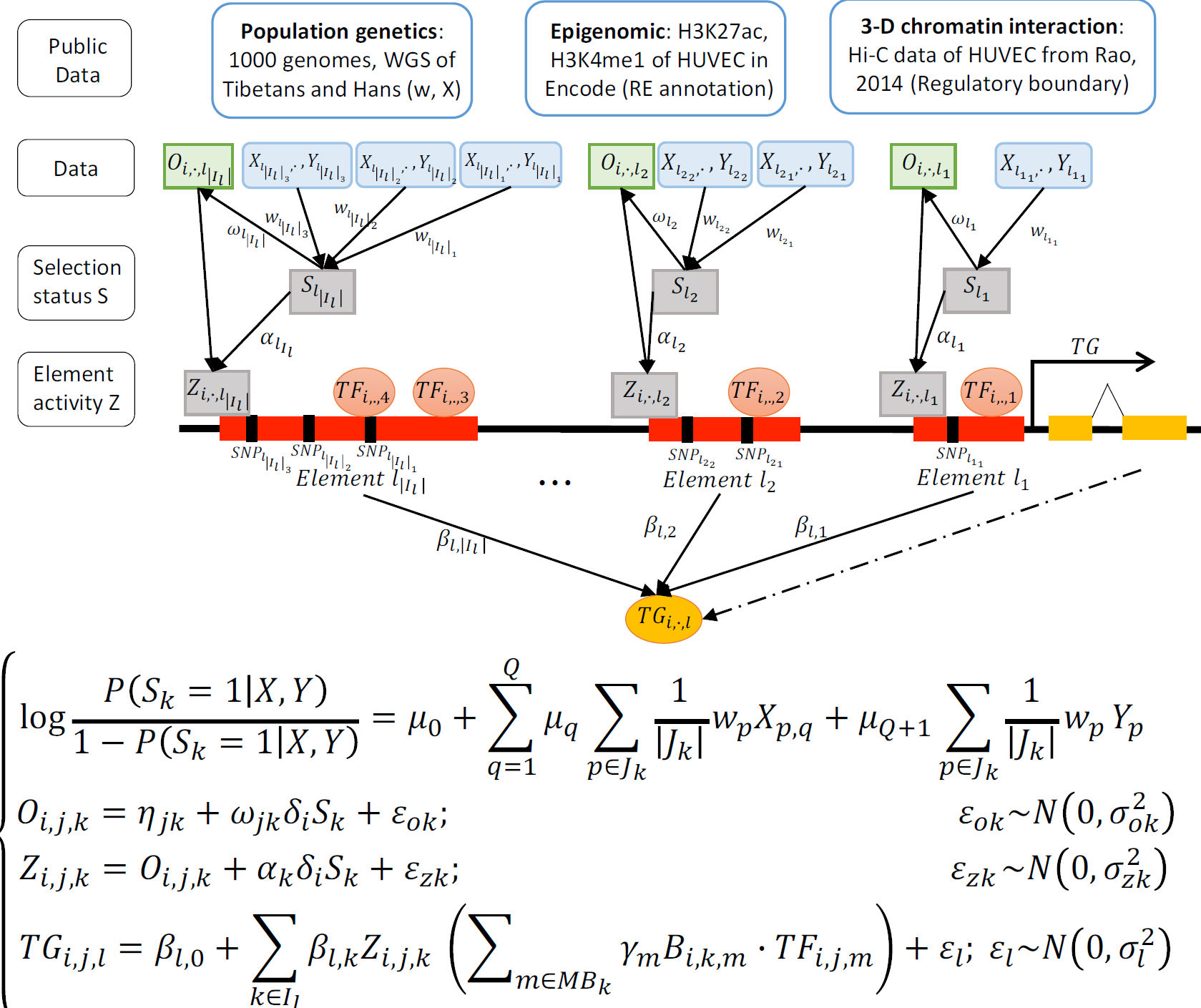
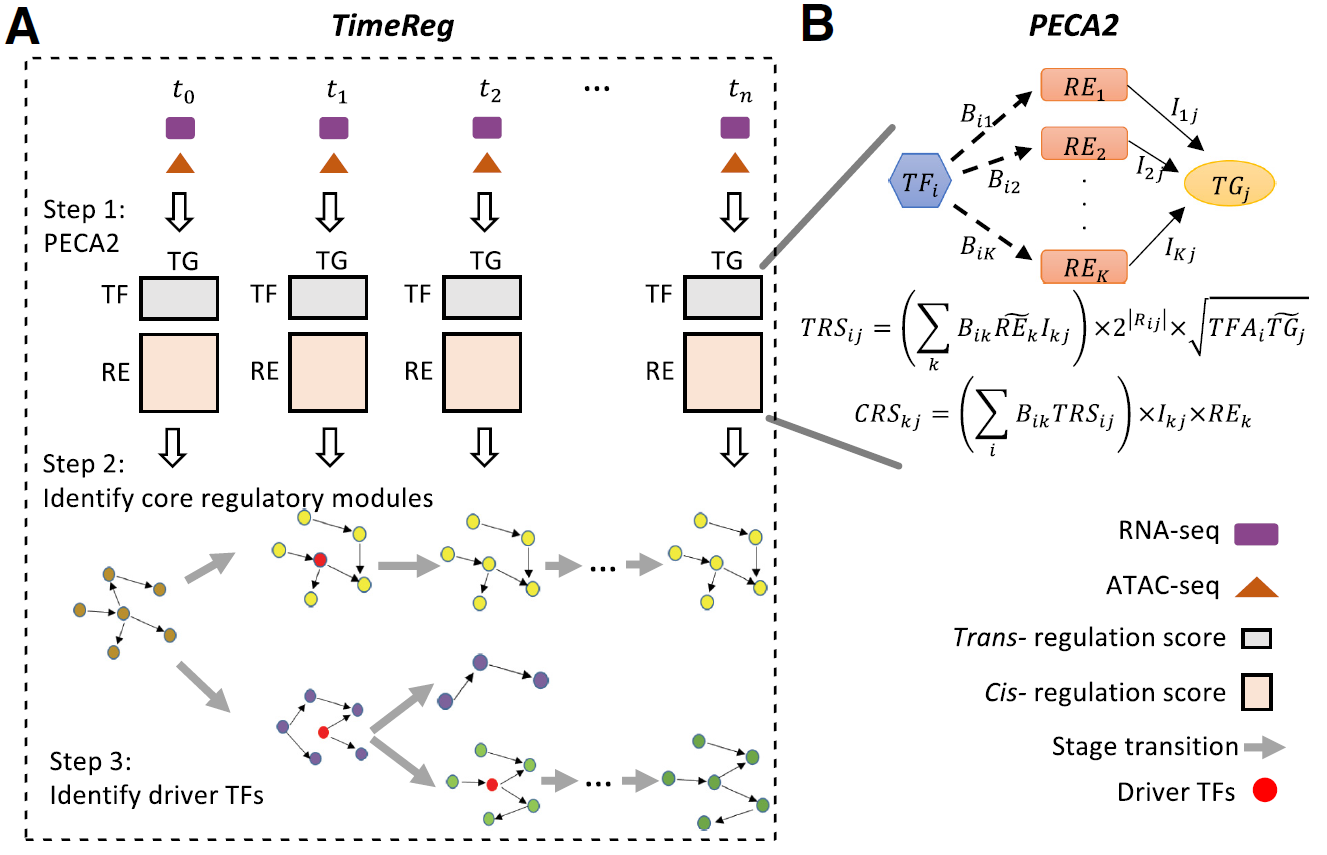
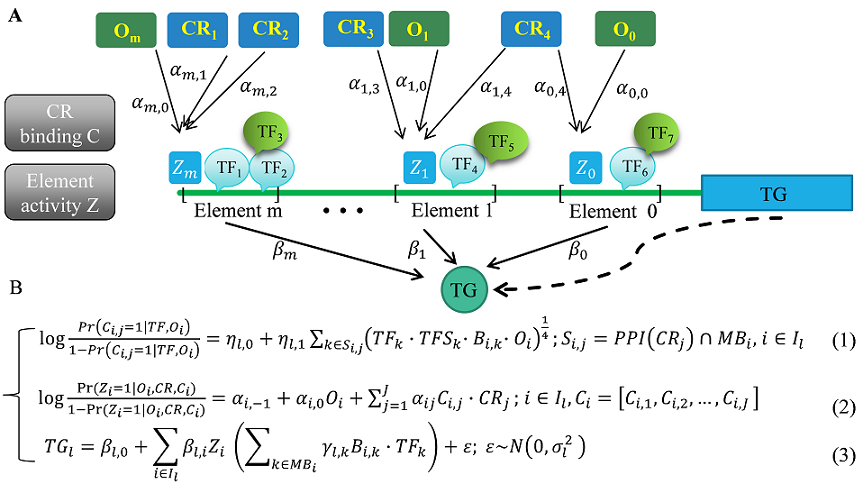
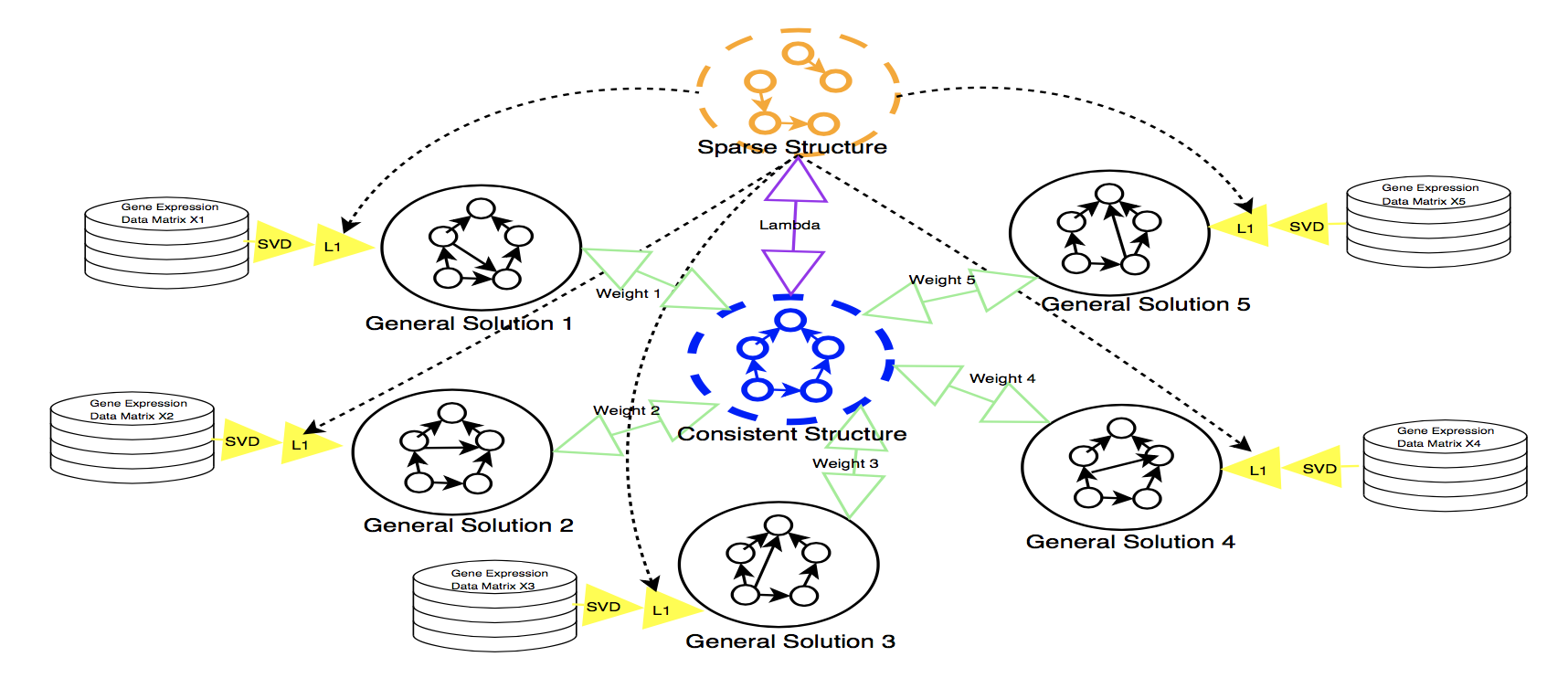
- SpecVar: a tool to reveal relevant tissue and associated the reulations among regulatory element and target gene for GWAS study.
- 3SCover: a method to identify safeguard TF from cell type -TF network by an extended mimimum set cover model.
- GuidingNet: a method to reveal transcriptional cofactor and predict binding for DNA methyltransferase by network regularization.
- scTIM: A convenient tool for marker detection based on single cell RNA-seq data.
- DC3: Deconvolution and coupled clustering from bulk and single cell genomics data
- CoupledNMF: Integrative analysis of single cell genomics data by coupled nonnegative matrix factorizations
- PECA2: a software for inferring context specific gene regulatory network from paired gene expression and chromatin accessibility data
- EllipsoidFN: a tool for identifying a heterogeneous set of cancer biomarkers based on gene expressions
- GNTInfer: A Gene Network reconstruction tool with compound Targets by Integrating multiple microarray datasets and prior information
- MILP_k: Mixed Integer Linear Programming for multiple-biomarker panel identification
- MNAligner: Alignment of molecular networks by quadratic programming
- Mimic: Mixed Integer programming Model to Identify Cell type specific marker panel
- NCCAUC: Nearest Centriod Classifier for AUC valuation
- NetPredATC: Network predicting drug's anatomical therapeutic chemical code
- PreDR: Drug repositioning from chemical, genomic and pharmacological data in an integrated framework
- PrePPItar: Machine learning framework to Predict PPI target for drug
- QMC: A combinatorial model and algorithm for globally searching community structure in complex networks
- NOA: Network Ontology Analysis
- DGPsubNet: Drug-Gene-Disease coherent SUBNETworks
- PRNA: Prediction of protein-RNA binding sites by a random forest method with combined features
- PSN: Predicting gene ontology functions from protein's regional surface structures
- SANA: protein Structure Alignment by Neighborhood Alignment
- Samo: a protein structure alignment tool based on multiple objective optimization
- InferPPI: Predicting protein interactions based on parsimony tendency of domain interactions
- MDCInfer: PPI prediction tool based on multiple domain cooperation analysis
LINKS
Yong Wang Lab | Academy of Mathematics and Systems Science | Chinese Academy of Sciences